Bitcoin for beginnersamit bhardwaj
40 comments
Que significa botar liquido por el ano
Linkages between genes never have been complete, but this is usual and it is result from the exchange of segments between homologous chromosomes. An exchange between homologous chromosomes, called crossing over, results in the recombination of genes in the pair of chromosomes.
Genetic analysis is the dissection of the structure and function of the genetic material. In classic genetic analysis, progeny from crosses between parents with different genetic characters are analyzed to -determine the frequency with which differing parental alleles are associated in new combinations.
Progeny showing the parental combinations of alleles are called parentals, and progeny showing nonparental combinations of alleles are called recombinants. The process by which the recombinants are produced is called genetic recombination.
Through testcrosses, we can determine which genes are linked to each other and can then construct a linkage map, or genetic map, of each chromosome. In meiosis, the precursor cells of the sperm or ova must multiply and at the same time reduce the number of chromosomes to one full set. Ttraits controlled by single, major Mendelian genes are isolated using genetic linkage maps created from crossing experiments.
Classic genetic mapping has provided information that is useful in many aspects of genetic analysis. For example, knowing the locations of genes on chromosomes has been useful in recombinant DNA research and in experiments directed toward understanding the DNA sequences in and around genes. The focus of mapping studies is on constructing genetic maps of genomes with the use of both gene markers and DNA markers.
In other words, it is an allele that marks a chromosome or a gene. Gene markers are alleles of the kind we have discussed to this point in the text. DNA markers are molecular markers-that is, DNA regions in the genome that differ sufficiently between individuals and thus can be detected by the molecular analysis of DNA.
The goal of genome-mapping studies is to generate high-resolution maps of the chromosomes. Such maps are useful for investigating genes and their functions.
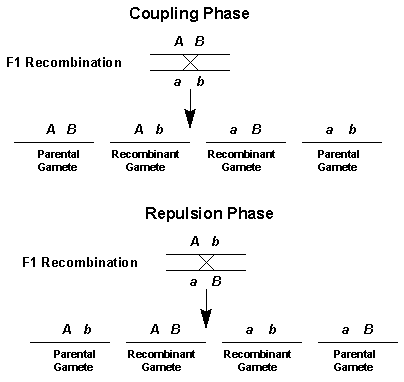
The ultimate genetic maps will be of the base-pair sequences of organisms' genomes. So if two genes are linked, the gametes produced during meiosis if no crossover occurs have only two allele combinations.
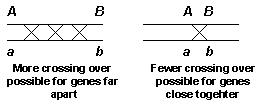
If a crossover occurs between the two linked genes, then the gametes produced have four allele combinations, just as if the genes assorted independently. The important question therefore becomes: The frequency of recombination between two points on a chromosome varies directly with the distance between the two points.
In other words, the closer together two genes are on a chromosome, the less likely a crossover is to occur between them. The farther they are apart, the more likely a crossover is to occur between them. If a crossover occurs randomly at one point on a chromosome, it's far more likely to occur between two genes if they're far apart than if they're close together. This property can be used to get an idea of the distance between two genes on a chromosome.
The more frequently a crossover event occurs between two genes, the farther apart they are. This is the basic concept behind gene mapping.
To put this another way, the genetic distance between two points on a chromosome equals the average number of crossovers between them. To express the genetic distance, we need to define a unit of measure: A map unit is sometimes referred to as a centiMorgan after T.
Morgan, who pioneered the use of Drosophila for the study of genetics. The document was created: Prepared under project CZ.